R for modellers - Vignette 08
Acquiring data
Julien Arino


Department of Mathematics
University of Manitoba*
* The University of Manitoba campuses are located on original lands of Anishinaabeg, Cree, Oji-Cree, Dakota and Dene peoples, and on the homeland of the Métis Nation.
Be “data aware”
Be data aware
- Using R (or Python), it is really easy to grab data from the web, e.g., from Open Data sources
- More and more locations have an open data policy
- As a modeller, you do not need to have data everywhere, but you should be aware of the context
- If you want your work to be useful, for instance in public health, you cannot be completely disconnected from reality
Data is everywhere
Closed data
- Often generated by companies, governments or research labs
- When available, come with multiple restrictions
Open data
- Often generated by the same entities but “liberated” after a certain period
- More and more frequent with governments/public entities
- Wide variety of licenses, so beware
- Wide variety of qualities, so beware
Open Data initiatives
Recent movement (5-10 years): governments (local or higher) create portals where data are centralised and published
Data gathering methods
- Example: population of South Africa
- Example - Dutch Elm Disease
- Data wrangling
- JA. Mathematical epidemiology in a data-rich world. Infectious Disease Modelling 5:161-188 (2020)
- See also GitHub repo for that paper
Data gathering methods
- By hand
- Using programs such as Engauge Digitizer or g3data
- Using natural language processing and other web scraping methods
- Using APIs
- Using R or Python packages (to interface with APIs)
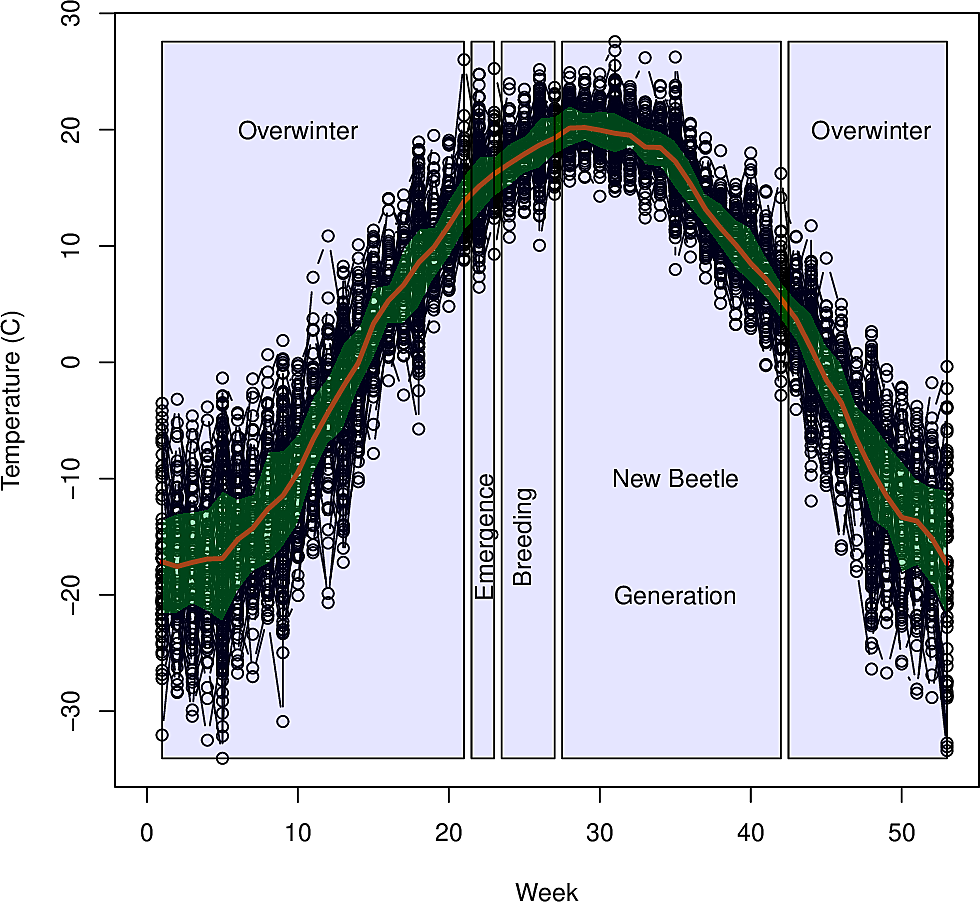
Example - Dutch Elm Disease
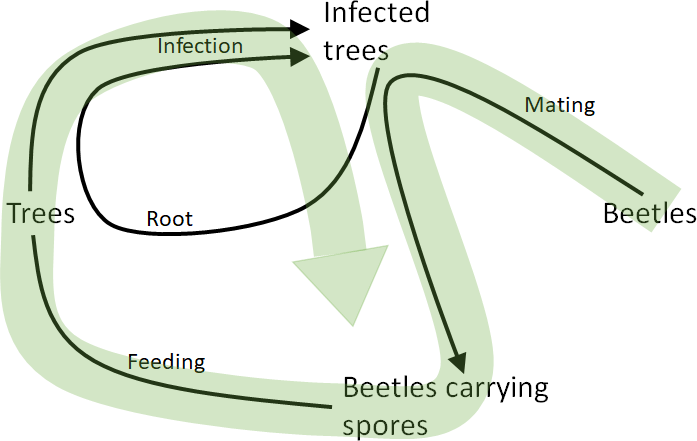
Dutch Elm Disease
- Fungal disease that affects Elms
- Caused by the fungus Ophiostoma ulmi
- Transmitted by the Elm bark beetle (Scolytus scolytus)
- Has decimated North American urban elm forests
bg contain
bg contain
bg contain
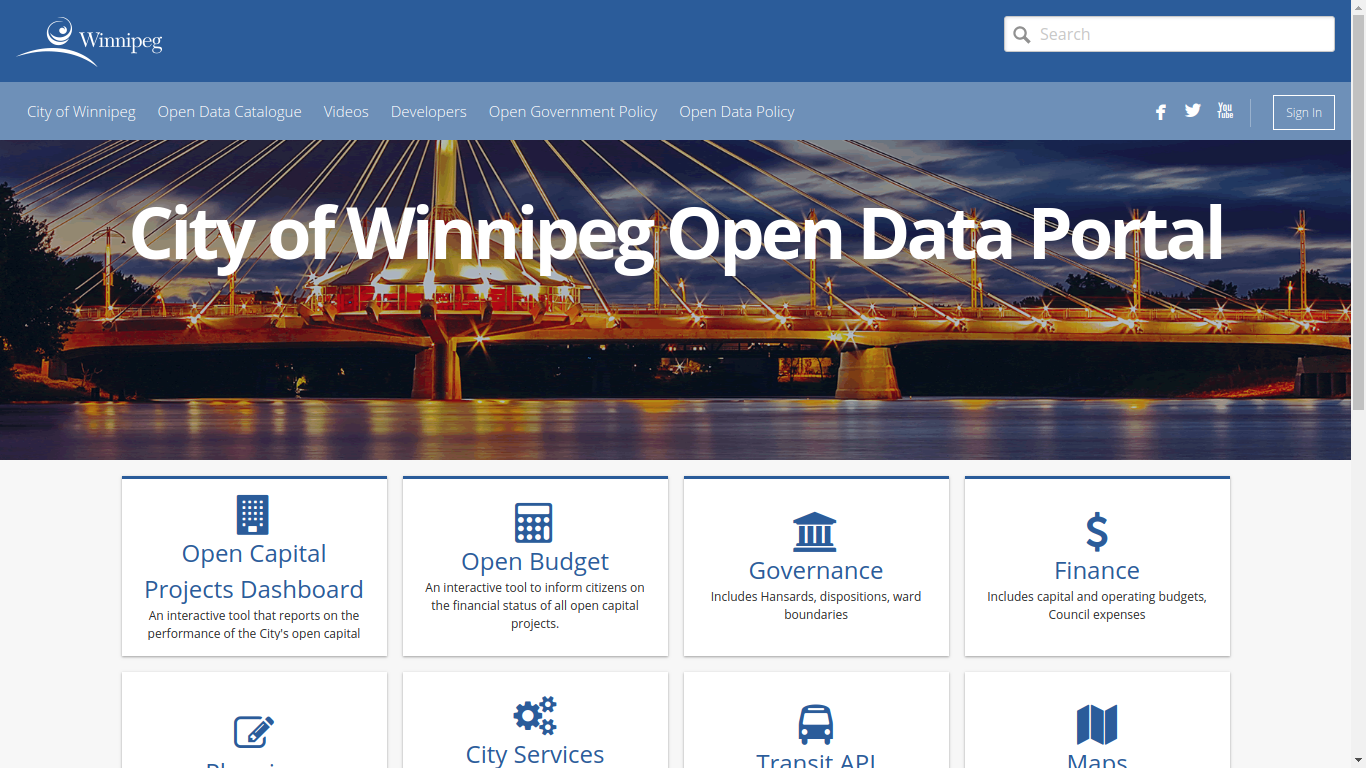
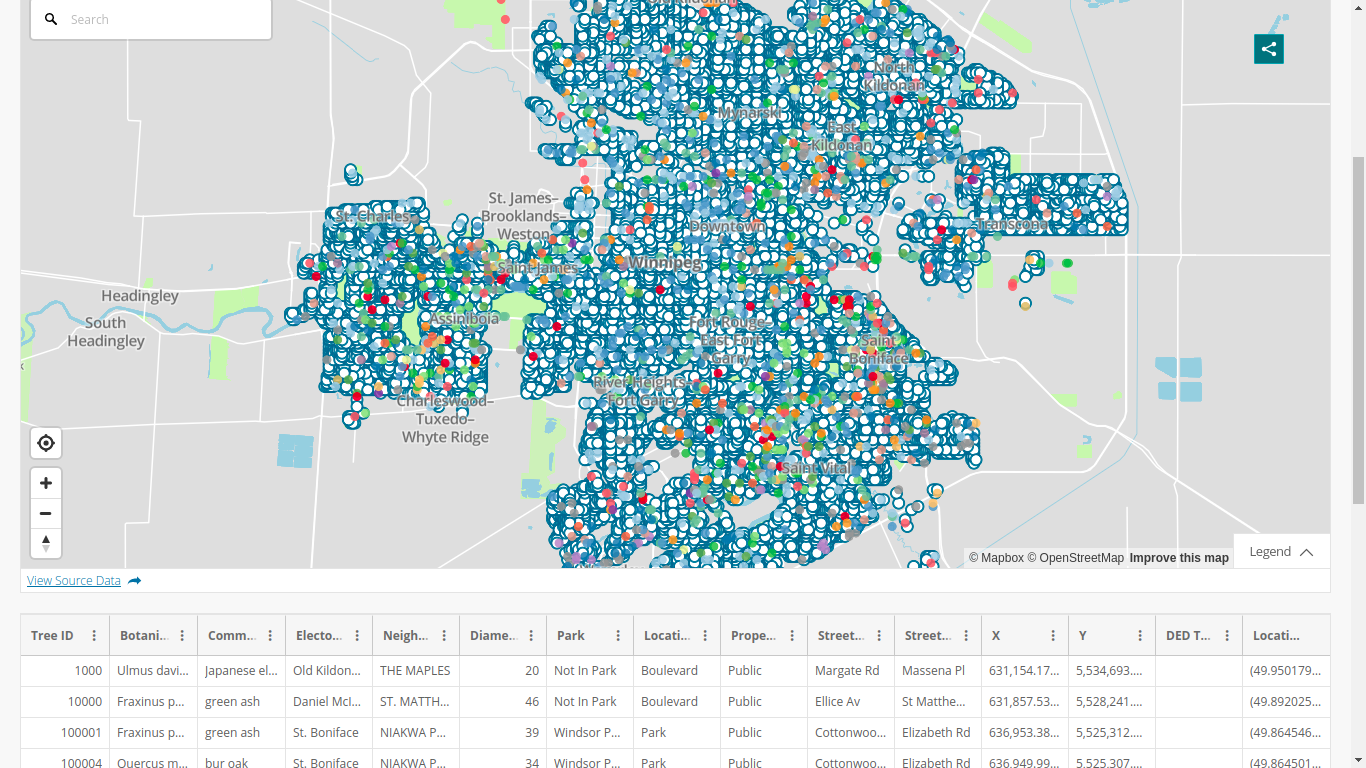
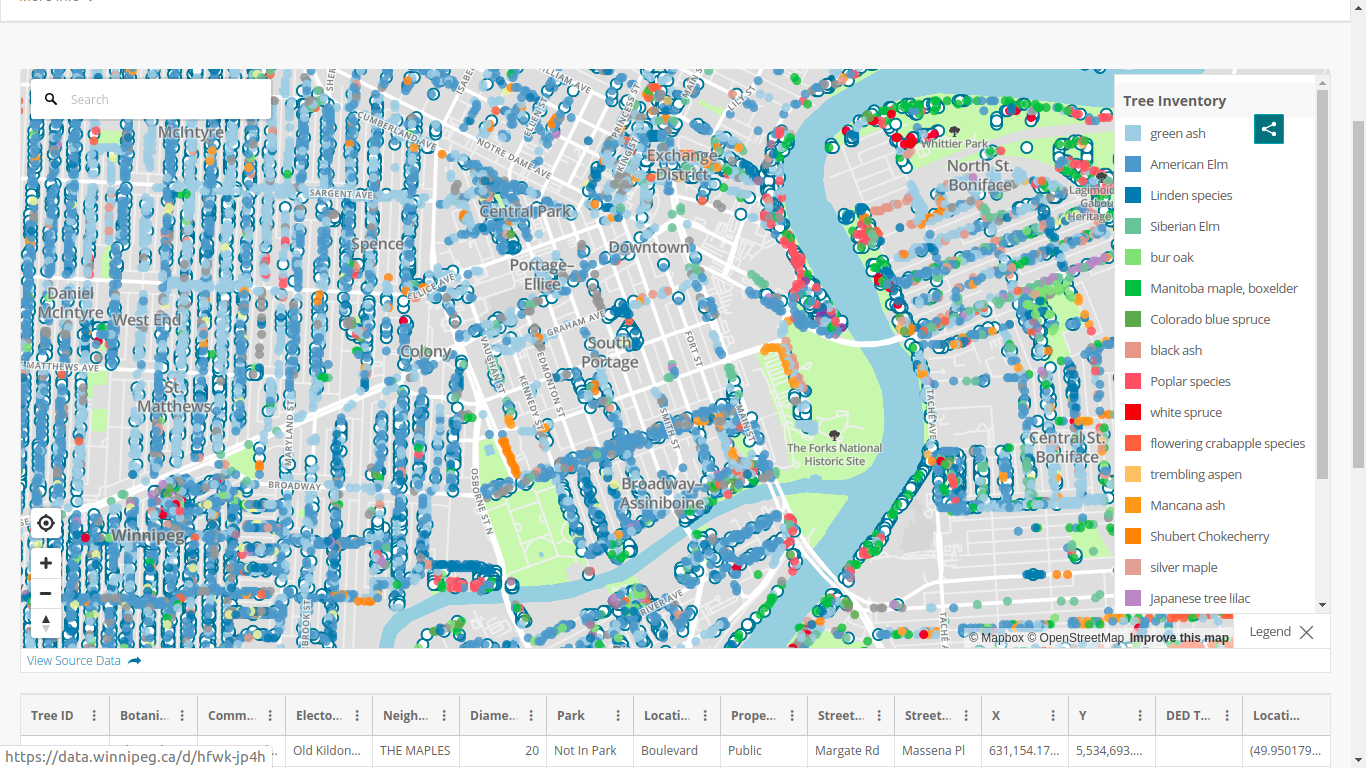
Getting the tree data
allTrees = read.csv("https://data.winnipeg.ca/api/views/hfwk-jp4h/roAfter this,
dim(allTrees)
## [1] 300846
15Let us clean things a little
elms_idx = grep("American Elm", allTrees$Common.Name, ignore.case = TRUE)
elms = allTrees[elms_idx, ]We are left with 54,036 American elms
bg contain
bg contain
Computation of root systems interactions
(Needs a relatively large machine here - about 50GB RAM)
- If roots of an infected tree touch roots of a susceptible tree, fungus is transmitted
- Spread of a tree’s root system depends on its height (we have diametre at breast height, DBH, for all trees)
- The way roadways are built, there cannot be contacts between root systems of trees on opposite sides of a street
Distances between all trees
elms_xy = cbind(elms$X, elms$Y)
D = dist(elms_xy)
idx_D = which(D<50)indices_LT is a large \(N(N-1)/2\times 2\) matrix with indices (orig,dest) of trees in the pairs of elms, so indices_LT[idx_D] are the pairs under consideration
Keep a little more..
indices_LT_kept = as.data.frame(cbind(indices_LT[idx_D,],
D[idx_D]))
colnames(indices_LT_kept) = c("i","j","dist")Create line segments between all pairs of trees
tree_locs_orig = cbind(elms_latlon$lon[indices_LT_kept$i],
elms_latlon$lat[indices_LT_kept$i])
tree_locs_dest = cbind(elms_latlon$lon[indices_LT_kept$j],
elms_latlon$lat[indices_LT_kept$j])
tree_pairs = do.call(
sf::st_sfc,
lapply(
1:nrow(tree_locs_orig),
function(i){
sf::st_linestring(
matrix(
c(tree_locs_orig[i,],
tree_locs_dest[i,]),
ncol=2,
byrow=TRUE)
)
}
)
)A bit of mapping
library(tidyverse)
# Get bounding polygon for Winnipeg
bb_poly = osmdata::getbb(place_name = "winnipeg",
format_out = "polygon")
# Get roads
roads <- osmdata::opq(bbox = bb_poly) %>%
osmdata::add_osm_feature(key = 'highway',
value = 'residential') %>%
osmdata::osmdata_sf () %>%
osmdata::trim_osmdata (bb_poly)
# Get rivers
rivers <- osmdata::opq(bbox = bb_poly) %>%
osmdata::add_osm_feature(key = 'waterway',
value = "river") %>%
osmdata::osmdata_sf () %>%
osmdata::trim_osmdata (bb_poly)And we finish easily
- We have the pairs of trees potentially in contact with each other
- We have the roads and rivers of the city, which is a collection of line segments
- If there is an intersection between a tree pair and a road/river, then we can forget this tree pair as their root systems cannot come into contact
st_crs(tree_pairs) = sf::st_crs(roads$osm_lines$geometry)
iroads = sf::st_intersects(x = roads$osm_lines$geometry,
y = tree_pairs)
irivers = sf::st_intersects(x = rivers$osm_lines$geometry,
y = tree_pairs)tree_pairs_roads_intersect = c()
for (i in 1:length(iroads)) {
if (length(iroads[[i]])>0) {
tree_pairs_roads_intersect = c(tree_pairs_roads_intersect,
iroads[[i]])
}
}
tree_pairs_roads_intersect = sort(tree_pairs_roads_intersect)
to_keep = 1:dim(tree_locs_orig)[1]
to_keep = setdiff(to_keep,tree_pairs_roads_intersect)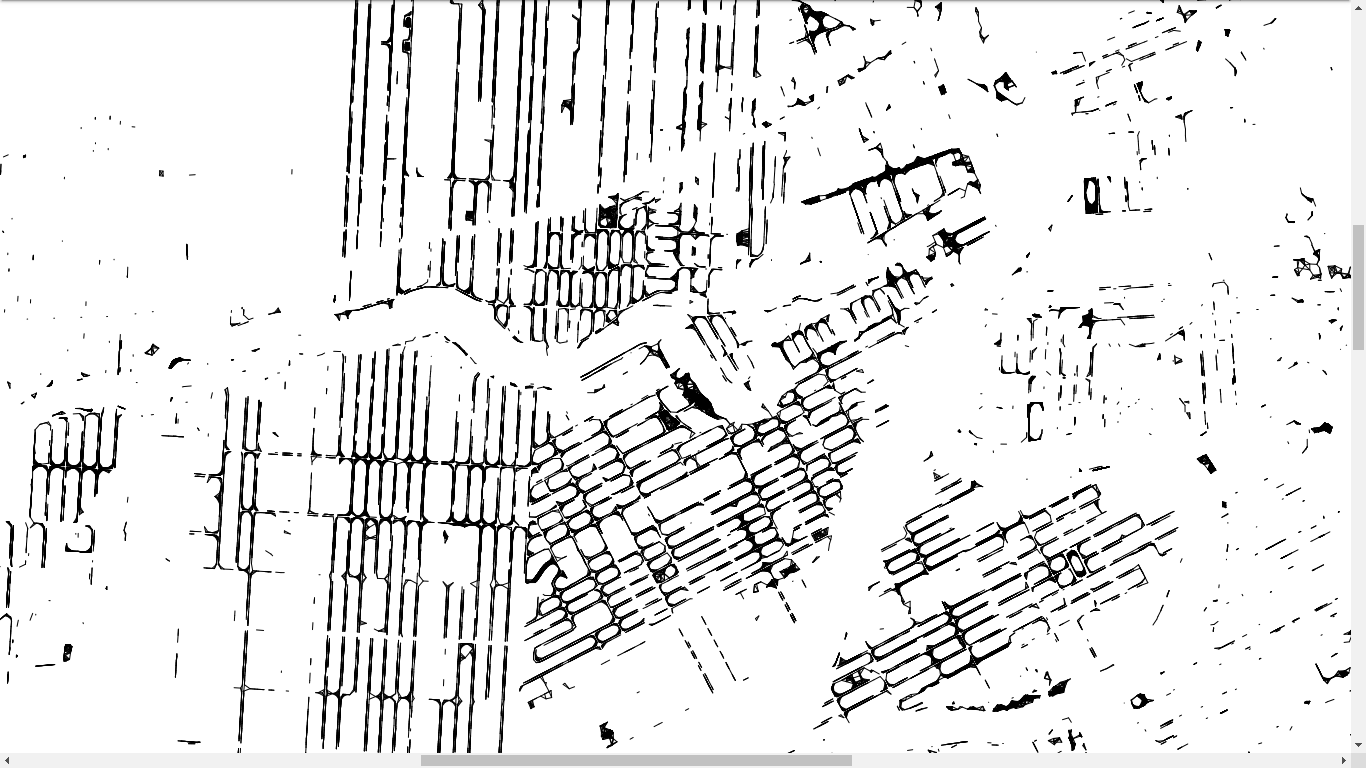
bg contain
Example: population of South Africa
library(wbstats)
pop_data_CTRY <- wb_data(country = "ZAF", indicator = "SP.POP.TOTL",
mrv = 100, return_wide = FALSE)
y_range = range(pop_data_CTRY$value)
y_axis <- make_y_axis(y_range)
png(file = "pop_ZAF.png",
width = 800, height = 400)
plot(pop_data_CTRY$date, pop_data_CTRY$value * y_axis$factor,
xlab = "Year", ylab = "Population", type = "b", lwd = 2,
yaxt = "n")
axis(2, at = y_axis$ticks, labels = y_axis$labels, las = 1)
dev.off()
crop_figure("pop_ZAF.png")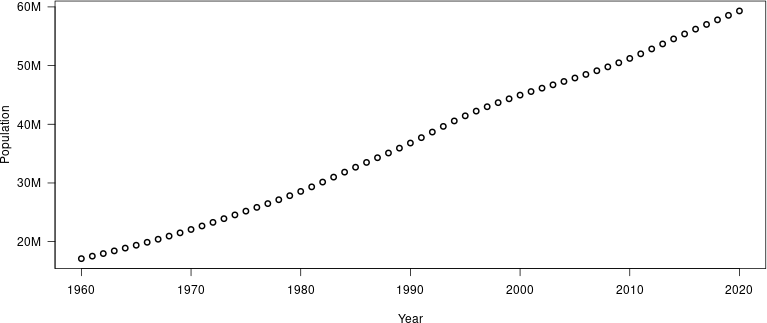
bg contain